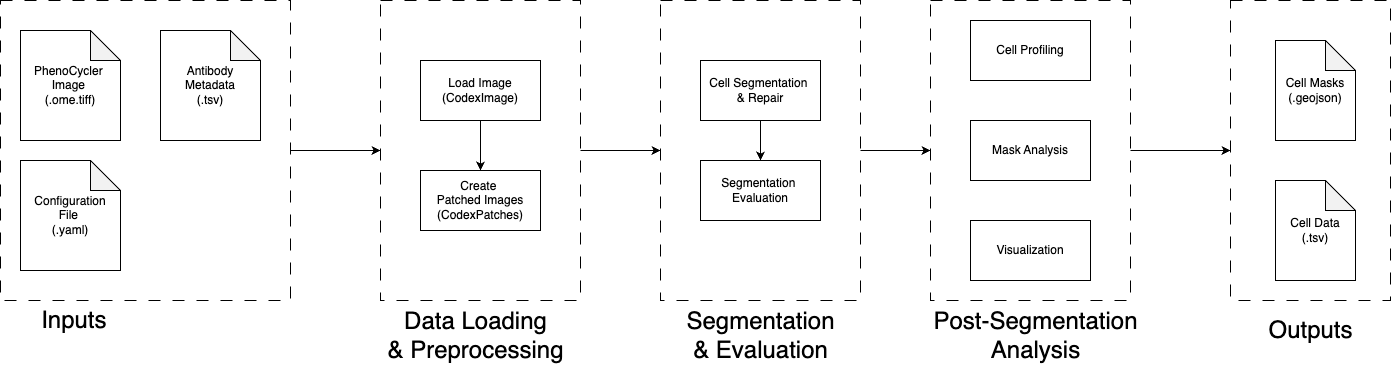
Overview
Aegle is a comprehensive pipeline for analyzing multiplex imaging data from PhenoCycler/CODEX platforms, providing end-to-end processing from raw images to single-cell insights.
Pipeline Architecture
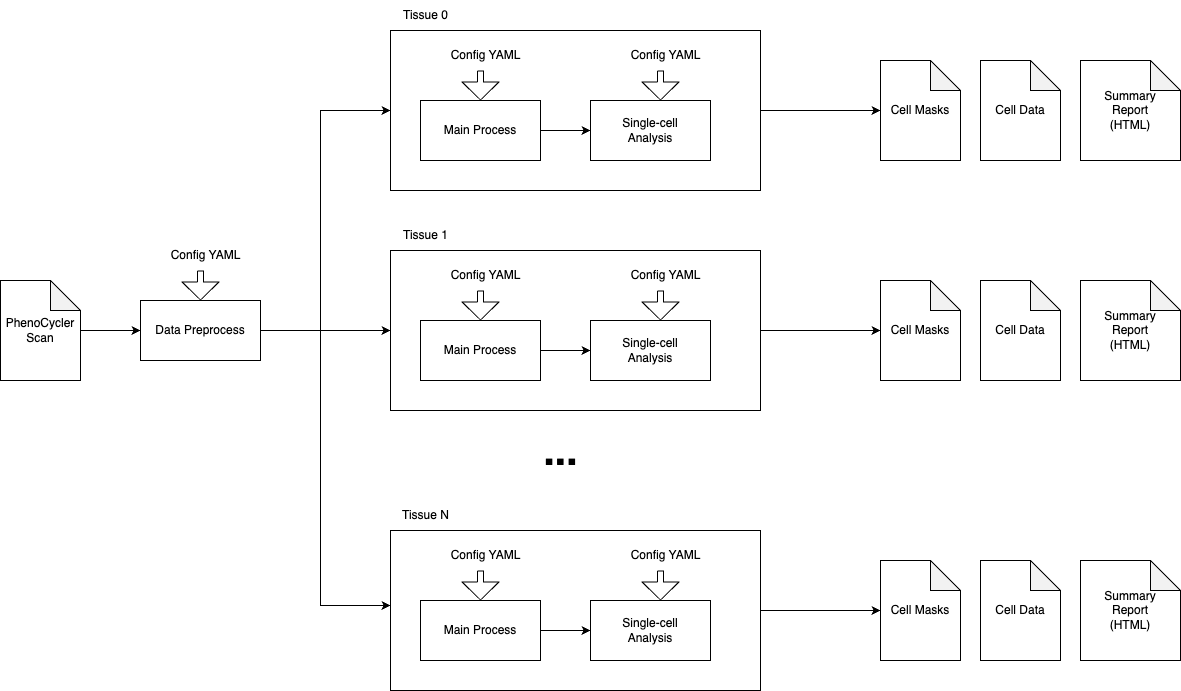
For detailed documentation, see the specific sections in the sidebar.
Experiment Configuration
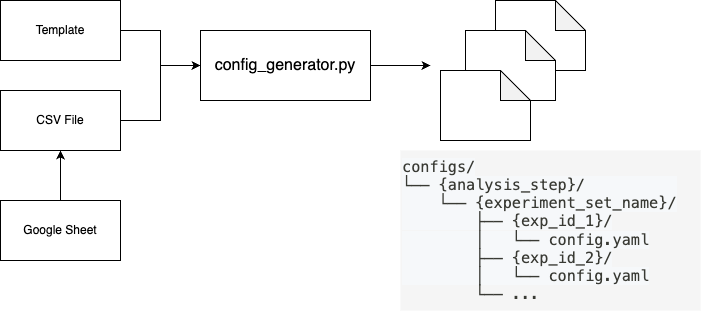
The configuration system automates experiment setup through:
- CSV Design Tables: Define experiments in Google Sheets and export to CSV
- YAML Templates: Standardized parameter templates for reproducibility
- Batch Generation: Generate configurations for multiple experiments simultaneously
- Validation: Automatic type conversion and parameter validation
Quick Start:
cd exps/
python config_generator.py
Data Preprocessing
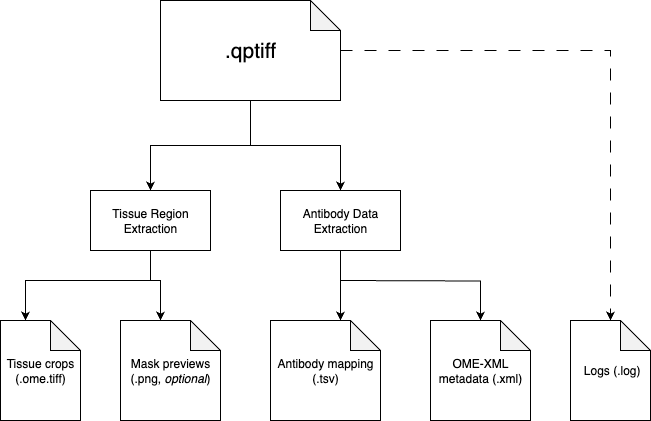
Preprocessing transforms raw QPTIFF files into analysis-ready data:
1. Tissue Extraction
- Manual annotation (recommended): Interactive napari-based tool
- Automatic detection: Computer vision-based tissue identification
2. Antibody Metadata
- Extracts channel-to-antibody mappings from QPTIFF metadata
- Generates standardized TSV files for downstream analysis
Execution:
./run_preprocess_ft.sh
Main Processing